
|
RATT: Rapid Annotation Transfer Tool |

|
RATT is software to transfer annotation from a reference (annotated) genome to an unannotated query genome.
NEW: RATT is not now part of PAGIT - Post Assembly Genome Improvement Toolkit. It is now easier to install and bundled with other usefull software.
It was first developed to transfer annotations between different genome assembly versions. However, it can also transfer annotations between strains and even different species, like Plasmodium chabaudi onto P. berghei, between different Leishmania species or Salmonella enterica onto other Salmonella serotypes. RATT is able to transfer any entries present on a reference sequence, such as the systematic id or an annotator's notes; such information would be lost in a de novo annotation.
It is important to notice, that the quality of the transferred annotation onto the query, depends on the quality of the annotation of the reference !!!
We are happy to annouce that RATT got accepted for publication:
Thomas D. Otto, Gary P. Dillon, Wim S. Degrave and Matthew Berriman, RATT: Rapid Annotation Transfer Tool, Nucleic Acid Research.
However, RATT checks whether gene models have changed between the two sequences and can correct changed start and stop codons, or frameshifts. The following figure outlines RATT's workflow:
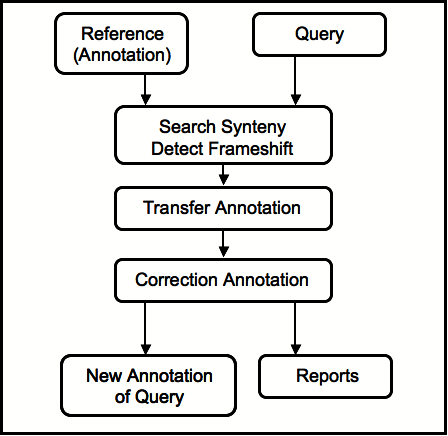
RATT returns the following statistics and information:
- How many tags are transferred correctly.
- Where CDSs different from the parent sequence.
- Which TAGs could not be transferred (usually due to indels or divergent sequence).
- New insertions in the query are shown, for further annotation.
RATT is rapid and assigns models with a high level of confidence. Nevertheless, genomes are not static entities and there will be changes between strains/species (often functionally informative). In these cases RATT will flag up all differences and report them to the annotator so they know where to focus their attention. The output of RATT can be best analyzed using Artemis and Act, see in the documentation.
Example
An example is given showing the transfer of the Mycobacterium tuberculosis strain H37Rv annotations onto the F11 strain. The data can be downloaded, run, or just visualized. Screenshots are shown.Download
To download RATT please use svn:
svn co "https://svn.code.sf.net/p/ratt/code/" ratt-code