|
RATT applied on tuberculosis |

|
Example
Here we show how RATT performs when transferring annotation between strains of Mycobacterium tuberculosis (H37Rv mapped to the F11 strain). The following is also intended to be a guide for new users using the H37Rv and F11 dataset. Screenshots and statistics are provided for evaluation purposes.Start the analysis
[ If you don't want to follow these steps, just download this. ]To try RATT, download the embl file for M. tuberculosis H37Rv and save it as Tb_H37Rv.embl in the directory embl. You will also need the fasta sequence for the F11 strain embl file (save as: F11.original.embl), which we have extracted here (save as: F11.fasta). Assuming that RATT is installed, it can be started by:
$RATT_HOME/start.ratt.sh embl F11.fasta F11 SimStrain
After less than 2 minutes the analysis should be complete and the following statistics shown:

Examining the results
RATT will generate several files, the most important are the F11.embl, F11.final.embl and F11.report.gff.- F11.embl (the uncorrected transfer)
- The F11.final holds the final (corrected) result.
- F11.report.gff records the differences between the annotation of the reference and the transferred annotation.
art F11.fasta + F11.final.embl + F11.report.gff.
Comparison view of results
To compare H37Rv with the F11 strain download this blast comparison file and start ACT as follows:act embl/Tb_H37Rv.embl comp.Tb.blast F11.fasta
Open the annotation files, by clicking on File -> F11.fasta -> open entries and select the files F11.final.embl and F11.report.gff.
One can see that the first gene models have transfered perfectly.
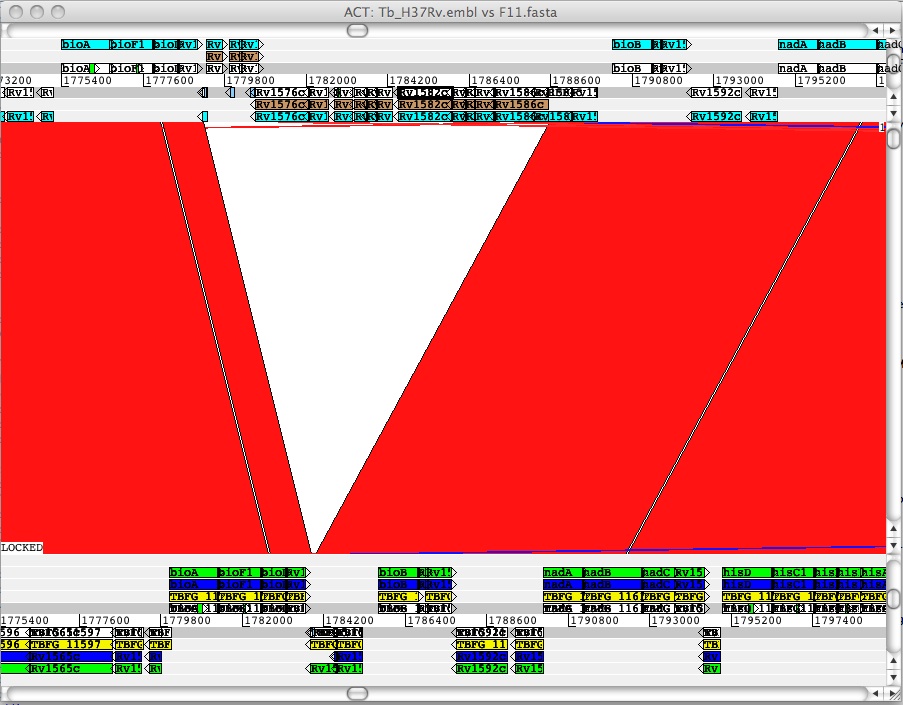
To see regions where the annotation couldn't be transferred, load the file F11.H37Rv.NOTtransfer.embl onto the Tb_H37Rv.embl file (Menu: File -> Tb_H37Rv.embl -> New Entry). For comparative purposes load the entries F11.orignal.embl and F11.embl onto the F11.fasta file (Menu: File -> Tb_H37Rv.embl -> New Entry). Next right mouse click over the F11 genome sequence, a pop-up will show: Select "one line per entry". Please repeat this for the H37Rv genome.
You should now see the following:
Figure:Mapping over a deletion. Due to a deletion initially the gene CAB09082 could not be transferred correctly (dark blue gene, below, in the middle). The correction step (green) has restored the CDS so it is equal to the F11 annotation in genbank (yellow). The gene models (brown) that cannot be mapped because they are inside a deletion are not transferred marked accordingly. NB Light blue denotes the orignal H37Rv gene models.
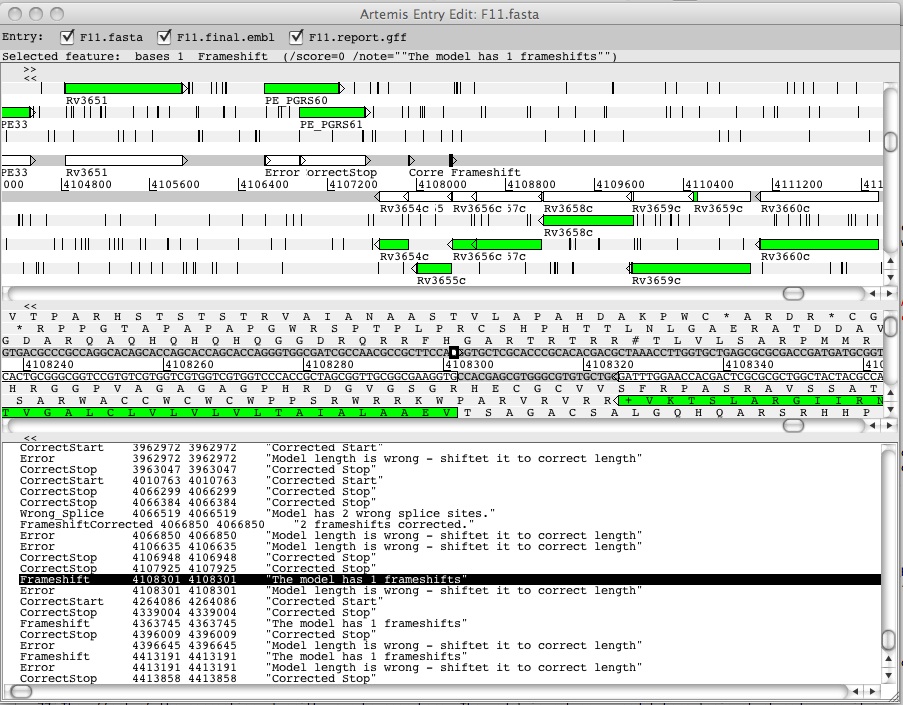
Correcting transferred models
The effect of the correction algorithm can be seen here. The transferred model does not possess an appropriate start codon, this is detected and corrected by RATT:
As can be seen, RATT is fast and produces precise results. Never-the-less, no annotation transfer will be perfect. Therefore, RATT will flag regions of concern for the user to manually examine. This way, an annotator can quickly assess the differences between strains, and thus take far less time manually checking annotations cf. gene finders.